2. 上海海洋大学水产与生命学院,上海 201306;
3. 广东省海洋生物种业工程技术研究中心,广东 广州 510300;
4. 广东海大集团股份有限公司,广东 广州 511400
Kisspeptin是由Kiss基因编码的神经多肽激素,在调控机体生长发育、能量代谢和生殖活动中发挥了重要作用[1]。Kiss基因存在Kiss1和Kiss2共2种类型。大多数哺乳动物仅有Kiss1基因,爬行类绿安乐蜥(Anolis carolinensis)中仅发现Kiss2的存在,鸟类中尚未发现Kiss基因的表达[2]。两栖类中非洲爪蟾(Xenopus tropicalis)有2种Kiss1基因,分别为Kiss1a和Kiss1b,同时存在Kiss2。硬骨鱼类有Kiss1和Kiss2共2种基因,但在进化过程中部分鱼类丢失了Kiss1基因,如青斑河鲀(Tetraodon nigroviridis)[3]、塞内加尔鳎(Solea senegalensis)[4]和三刺鱼(Gasterosteus aculeatus)[5]。Kiss基因在不同脊椎动物中存在的类型差异可能是由于脊椎动物进化过程中基因组的复制和长期自然选择的结果[6-7]。基因组复制导致基因类型增多,而长期的自然选择使得一些功能冗杂的序列在进化中丢失[8]。至今已在青鳉(Oryzias latipes)[9-10]、斑马鱼(Danio rerio)[10-11]、花鲈(Lateolabrax japonicus)[5,12]、金鱼(Carassius auratus)[13-14]、日本鲭(Scomber japonicas)[15]、条纹鲈(Morone saxatilis)[16]、红鳍东方鲀(Takifugu rubripes)、青斑河鲀[12]、星点东方鲀(T. niphobles)[11]、斜带石斑鱼(Epinephelus coioides)[17]、塞内加尔鳎[18]及三刺鱼[3]等硬骨鱼中克隆到Kiss。目前关于卵形鲳鲹Kiss的研究未见报道。
Kiss1系统在能量代谢和机体生长中发挥重要作用[12,19],维持机体葡萄糖稳态的相关激素如胰岛素可影响Kiss1的表达,小鼠Kiss1神经元上的胰岛素受体缺失,会使Kiss1表达水平下降[20]。短期禁食可显著提升小鼠脂肪组织中Kiss1 mRNA的水平,可以促进能量形式的转变[21]。代谢相关激素瘦素(leptin, LP)和神经肽Y (neuropeptide Y, NPY)均可刺激小鼠离体下丘脑细胞系N6中的Kiss1 mRNA上调表达,说明下丘脑Kiss1系统参与机体能量代谢调节[22]。kisspeptin也可影响生长相关激素如生长激素(growth hormone, GH)和生长乳素(somatolactin α, SLα)的释放。用kisspeptin多肽处理金鱼体外垂体细胞可使生长激素(GH)的转录表达升高[23]。金鱼kisspeptin多肽可以通过cAMP/PKA和磷脂酶C (PLC)/PKC途径以及随后的Ca2+/钙调蛋白(CaM)等信号通路调节垂体SLα的释放[23]。综上所述,Kiss1系统影响机体能量消耗和葡萄糖稳态[24],在代谢和生长方面起重要作用。
卵形鲳鲹(Trachinotus ovatus)隶属于硬骨鱼纲(Osteichthyes)、鲈形目(Perciformes)、鲹科(Carangidae)、鲳鲹属(Trachinotus),俗称金鲳、黄腊鲳[25]。它广泛分布在东南亚和地中海的热带和亚热带海域[26],包括中国的广东、广西、海南和福建沿海。由于其肉质鲜美、生长迅速,已成为华南沿海最重要的海水养殖鱼类之一[27]。Kiss1作为一种多功能激素在哺乳动物中被广泛研究,但在硬骨鱼中仅有生殖调控方面的研究,对于摄食调控生理和生长方面的研究未见报道。本实验拟在本课题组前期开展的颗粒饲料、鱿鱼以及杂鱼3种类型饵料投喂卵形鲳鲹实验[28]的基础上,研究卵形鲳鲹摄食3种不同饵料条件下Kiss1基因的表达模式,以期了解Kiss1基因在卵形鲳鲹摄食调控生理和生长方面的作用,为进一步挖掘其在硬骨鱼生殖以外的其他功能奠定基础。
1 材料与方法 1.1 实验材料实验所用卵形鲳鲹组织样品来源于本课题组Sun等[28]饲养的卵形鲳鲹,并用前期实验的3种饵料类型喂养,该实验在中国水产科学研究院海南陵水基地开展。取–80 °C保存的实验组的肝脏和肠道组织样品用于mRNA的提取。
为研究基因的组织表达谱,另取其他网箱养殖健康成年鱼(喂养实验外)3条,取心脏、肾脏、鳃、鳍、脾脏、眼睛、皮肤、肌肉、胃、肠道、脑和肝脏12个组织样品保存在RNAlater (Ambion, CA, USA)中并储存在–80 °C直到RNA提取。
1.2 总RNA和DNA的提取根据TRIzol说明书(TaKaRa,大连)提取卵形鲳鲹实验组的肝脏、肠道组织及健康卵形鲳鲹12个组织的RNA。对RNA进行检测(用1%琼脂糖凝胶电泳)并测定RNA的OD值。RNA各取2 μg合成cDNA (使用TaKaRa的PrimeScript RT reagent Kit with gDNA Eraser试剂盒)用于qRT-PCR。健康卵形鲳鲹RNA取3 μg合成3′RACE cDNA (用TaKaRa试剂盒3′-Full RACE Core Set with PrimeScriptTM RTase)用于扩增3′末端全长序列。将cDNA样品储存在–20 °C。
1.3 基因克隆和测序卵形鲳鲹Kiss1基因序列在实验室通过基因组测序,组装和注释获得欧洲核酸档案库(European Nucleotide Archive, ENA)的登录号PRJEB22654;序列在BioProject PRJNA406847读取存档。卵形鲳鲹Kiss1基因cDNA序列通过随机测序技术获得,其中一个序列与GenBank数据库中其他硬骨鱼的Kiss1 mRNA, complete cds具有较高同源性,命名为ToKiss1,为了验证序列的准确性,用Primer Premier 5.0软件设计引物对ToKiss1-F/R (表1)进行PCR验证。ToKiss1开放阅读框序列,设计3′RACE引物ToKiss1-3′-GPS1/2,以卵形鲳鲹3′RACE cDNA为模板根据试剂盒说明书扩增ToKiss1基因3′端片段,对目的条带进行纯化,并与载体pMD18-T (TaKaRa,大连)进行连接,转化后进行阳性克隆筛选并测序(Invitrogen,广州)。
| 表 1 实验所用引物 Tab.1 Primers used in this study |
通过ProtParam (http://web.expasy.org/compute_pi/)检测分子量和pI值,并使用SignalP 4.1(http://www.cbs.dtu.dk/services/)进行信号肽预测。使用ExPASy程序(http://prosite.expasy.org/)、NetNGlyc程序(http://www.cbs.dtu.dk/services/NetNGlyc/)和Kinasephos程序(http://kinasephos.mbc.nctu.edu.tw/)对ToKiss1氨基酸序列上信号肽、糖基化位点及蛋白磷酸化位点等功能基序进行预测,分析ToKiss1功能位点保守性及结构作用;使用Expasy翻译工具(http://www.expasy.org)推断ToKiss1氨基酸序列。利用GOR方法(https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_phd.html)进行蛋白二级结构预测,同时使用SWISS-MODEL软件(http://swissmodel.expasy.org/)进行三级结构预测;通过使用Clustalw2程序(http://www.ebi.ac.uk/Tools/msa/clustalw2/)进行多重序列比对;其他物种Kiss1基因(表2)从NCBI数据库(https://www.ncbi.nlm.nih.gov/)中下载,使用MEGA 7.0软件用邻接法(neighbour-joining, NJ)、最大似然法(maximum likelihood, ML)和最大简约法(maximum parsimony, MP) 分别构建系统进化树,及对Kiss1与其他物种的氨基酸同源性和相似性进行比对分析。
| 表 2 进化树构建所用物种 Tab.2 Species applied to reconstruction of phylogenetic relationship |
根据ToKiss1基因cDNA序列设计特异性引物为To Kiss1-qF和 To Kiss1-qR,内参基因EF-1α基因的引物为EF-1α-F和EF-1α-R (表1)。将cDNA稀释为(60±5) ng/μL后作为荧光定量模板,按照TaKaRa SYBR PremixExtaqTM Kit (Perfect Real Time)试剂盒进行PCR反应,每个样品进行3次重复。实验结果采用相对荧光定量法(2–ΔΔCt法)分析ToKiss1基因在卵形鲳鲹各个组织及投喂实验中肝脏和肠道组织的表达量。
2 结果 2.1 ToKiss1基因cDNA克隆及生物信息学分析利用卵形鲳鲹转录组中Kiss1序列片段,通过克隆获得Kiss1基因cDNA序列(图1),命名为ToKiss1(GenBank登录号:MG843840)。ToKiss1 cDNA全长505 bp,5′非编码区(UTR)123 bp、开放阅读框(ORF)312 bp、3′非编码区(UTR)70 bp。ORF可编码104个氨基酸,理论分子量约为11.49 ku,pI为10.45。该序列包含1个N端信号肽(1~19 aa),13个磷酸化位点(其中丝氨酸位点7个、苏氨酸位点5个、酪氨酸位点1个),未发现糖基化位点。ToKiss1氨基酸序列与其他硬骨鱼Kiss1氨基酸序列相似,具有1个典型的kisspeptin-10结构域(92~101 aa)。

|
图 1 卵形鲳鲹Kiss1基因的cDNA序列和氨基酸序列 起始密码子(ATG)和终止密码子(TGA)用虚线框标出;信号肽序列用实线框标出;无规则卷曲结构域用阴影标出;双下划线标注为kisspeptin-10核心序列;单下划线标注为酪氨酸磷酸化位点;丝氨酸磷酸化位点加粗标出;苏氨酸磷酸化位点用圆圈标出;poly (A)加尾信号斜体加粗标出 Fig. 1 Full-length cDNA and amino acid sequences of ToKiss1 Virtual frame indicates initiation codon (ATG) and stop codon (TGA); solid frame indicates signal peptides; the low complexity domains are in grey shadow; the kisspeptin-10 signature sequence is double underlined; Cys residues are single underlined; Ser residues are in blod; Thr residues are circled; the poly (A) signal sequence is in italics and bold |
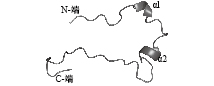
利用GOR法对ToKiss1蛋白质二级结构预测后发现,整条氨基酸序列的二级结构由40.78% α螺旋、16.50%延伸链以及42.72%无规则卷曲构成。SWISS-MODEL结果显示(图2),ToKiss1的三级结构由2个α螺旋以及连接的无规则卷曲构成。
|
图 2 卵形鲳鲹ToKiss1蛋白三级结构的模型 Fig. 2 Protein tertiary structure model of ToKiss1 |
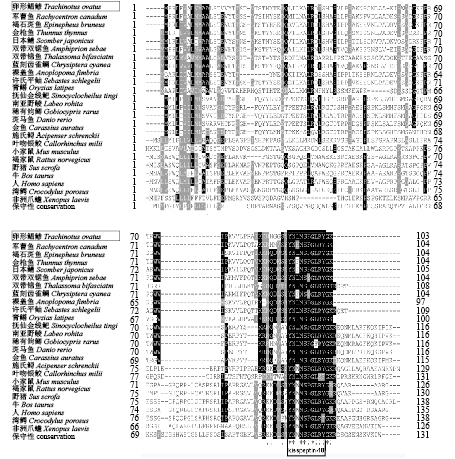
利用Clustal X软件对表2中的物种的氨基酸序列进行多重序列比对分析(图3),结果表明ToKiss1与其他物种的Kiss1氨基酸序列一样,具有保守的kisspeptin-10核心序列。哺乳动物的kisspeptin-10核心序列为“YNWNSFGLRY”;硬骨鱼的kisspeptin-10核心序列多为“YNLNSFGLRY”;软骨鱼叶吻银鲛的kisspeptin-10核心序列为“YNLNSFGLKF”(图3)。不同哺乳动物和硬骨鱼中均有个别氨基酸的差异,如硬骨鱼中金枪鱼、日本鲭、蓝刻齿雀鲷的第3位氨基酸为苯丙氨酸,施氏鲟(Acipenser schrenckii)的为色氨酸,军曹鱼的为蛋氨酸。双带双锯鱼和蓝刻齿雀鲷的第6位氨基酸为酪氨酸。稀有鮈鲫的第8位氨基酸为苯丙氨酸。哺乳动物中人的第10位氨基酸为苯丙氨酸。ToKiss1的氨基酸序列与其他硬骨鱼类、哺乳类、两栖类、爬行类的同源性分别为24.0%~75.9%、16.8~21%、14.8%和13.4% (表2)。ToKiss1的氨基酸序列与其同亚目的军曹鱼同源性最高(75.9%),与硬骨鱼中鲤形目的同源性最低(24%~26.9%)。卵形鲳鲹与其他鲈形目、鲉形目(Scorpaeniformes)、颌针鱼目等鱼类Kiss1氨基酸的两两之间的泊松校正距离较近,为0.202 2~0.895 4,与鲤形目的距离较远,为1.127 2~1.218 2。ToKiss1氨基酸与软骨鱼类、哺乳类、两栖类、爬行类之间存在更远的成对距离,其范围分别为1.490 1、1.218 2~1.429 5、1.490 1和1.623 6。
|
图 3 ToKiss1与其他物种的Kiss1氨基酸序列比对 图中基因、物种名及对应的登录号见表2,下同 Fig. 3 Multiple sequence alignment of amino acid sequence of ToKiss1 with Kiss1 from other organisms The genes, Latin names and their accession no. are shown in Tab. 2, the same below |
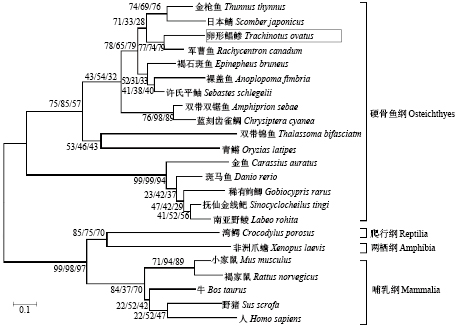
通过NJ、ML、MP 3种方法构建的系统进化树具有相似的聚类结果(图4)。硬骨鱼单独聚为一支。在这一大支上鲤形目单独聚为一支,鲈形目、鲉形目和颌针鱼目聚为一支,卵形鲳鲹与同亚目的军曹鱼亲缘关系最近。哺乳类、两栖类和爬行类聚为一支且与鱼类亲缘关系较远(图4)。
|
图 4 基于Kiss1基因编码的氨基酸序列构建的NJ/ML/MP系统进化树 Fig. 4 NJ/ML/MP phylogenetic tree based on amino acid sequences of Kiss1 gene |
ToKiss1基因(GenBank登录号:MG843839)全长2 768 bp,由3个外显子2个内含子组成,内含子和外显子接头遵循GT-AG规则(图5)。外显子1包括98 bp的非编码序列,外显子2包括25 bp的非编码序列和91 bp的编码序列,外显子3包括221 bp的编码序列和70 bp的非编码序列。内含子1和2分别为1 513和750 bp。

|
图 5 ToKiss1基因序列及编码的氨基酸序列 poly (A)加尾信号使用斜体加粗表示;内含子接头(gt/ag)使用矩形框表示;灰色阴影表示外显子序列 Fig. 5 Structure of the ToKiss1 genome and the deduced amino acid sequence The poly (A) signal sequence is in italics and bold; the introns splicing consensus (gt/ag) are in box; the exons are in grey shadow |
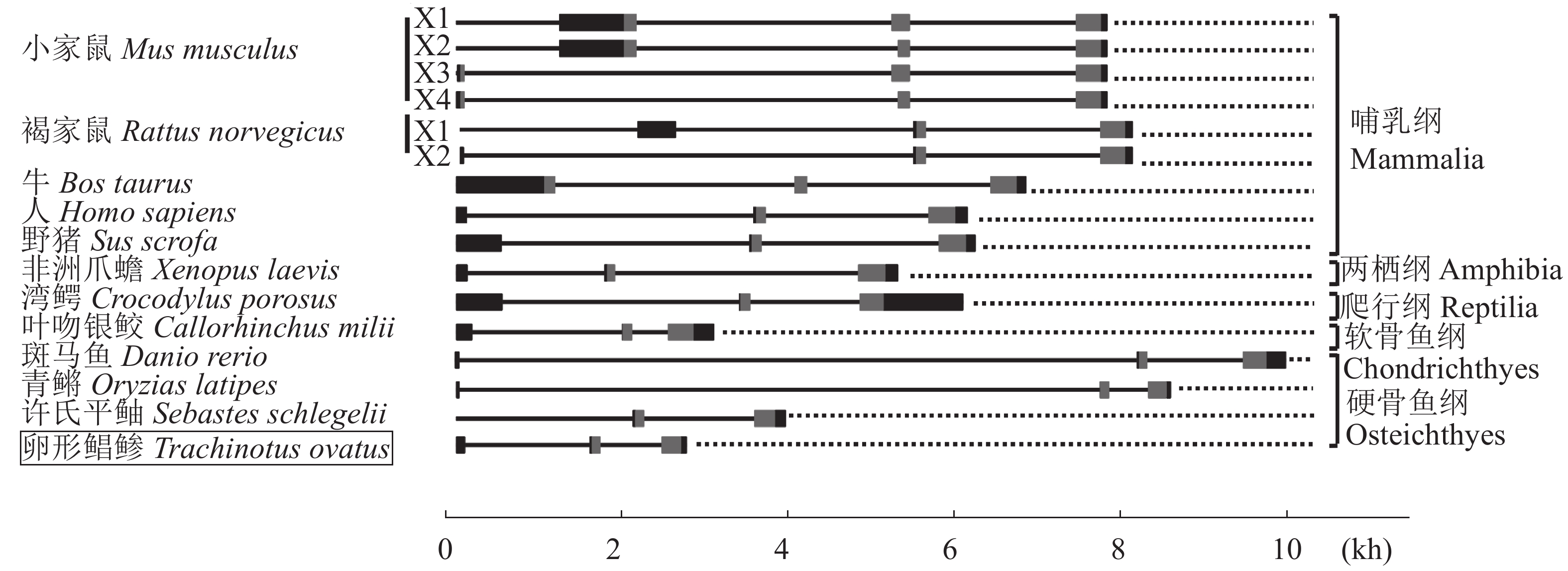
不同物种的Kiss1基因序列的内含子/外显子结构比较结果表明,所有脊椎动物的Kiss1基因均含有3个外显子和2个内含子,其中家鼠和牛Kiss1蛋白由外显子1、2和3编码,其他脊椎动物Kiss1蛋白由外显子2和3编码(图6)。此外,家鼠Kiss1外显子1和2均具有2种可变剪接体。物种间Kiss1基因长度差别较大,斑马鱼的最长,约为10 kb,卵形鲳鲹的最短,约为3 kb,主要是由内含子1的长度差别较大所导致。
|
图 6 Kiss1基因的外显子—内含子结构比较 矩形和直线分别代表外显子和内含子;灰色和黑色的矩形分别代表CDS和UTR;图中数字代表碱基对数;许氏平鲉Kiss1基因序列不完整(CDS完整);每个物种的Kiss1基因序列号:小家鼠(NC_000067.6);褐家鼠(NC_005112.4);牛(NC_037343.1);人(NG_032151.1);野猪(NC_010451.4);非洲爪蟾(NC_030727.1);湾鳄(NW_017728935.1);叶吻银鲛(NW_006890402.1);斑马鱼(NC_007122.7);青鳉(NC_019863.1);许氏平鲉(KJ139960.1) Fig. 6 Comparison of the exon–intron organization of the Kiss1 genes The boxes and bars represent the exons and introns, respectively. The boxes in gray and black represent the CDS and UTR, respectively. The numbers are the base pairs. The Kiss1 gene of S. schlegelii is incomplete (complete CDS). The Kiss1 sequence no. of each species: M. musculus (NC_000067.6); R. norvegicus (NC_005112.4); B. taurus (NC_037343.1); H. sapiens (NG_032151.1); S. scrofa (NC_010451.4); X. laevis (NC_030727.1); C. porosus (NW_017728935.1); C. milii (NW_006890402.1); D. rerio (NC_007122.7); O. latipes (NC_019863.1); S. schlegelii (KJ139960.1) |
利用qRT-PCR分析ToKiss1基因mRNA在正常卵形鲳鲹12个组织中的表达差异,结果显示ToKiss1基因mRNA在脑中的表达量最高,其次是肠道、胃、脾脏、肌肉和心脏,在肝脏、肾脏、鳃、鳍、眼睛和皮肤组织中表达量最低(图7)。
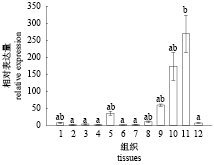
|
图 7 卵形鲳鲹ToKiss1基因mRNA在不同组织中的表达 1. 心脏,2. 肾脏,3. 鳃,4. 鳍,5. 脾脏,6. 眼睛,7. 皮肤,8. 肌肉,9. 胃,10. 肠,11. 脑,12. 肝脏;垂直线表示平均值±标准误(n=3),不同的字母表示差异显著(P<0.05),下同 Fig. 7 Tissues expression analysis of ToKiss1 mRNA 1. heart, 2. kidney, 3. gill, 4. fin, 5. spleen, 6. eye, 7. skin, 8. muscle, 9. stomach, 10. intestine, 11. brain, 12. liver; vertical bars represent mean±SE (n=3), the different letters mean significant difference (P>0.05), the same below |
利用qRT-PCR分析ToKiss1基因mRNA在不同饵料投喂实验组卵形鲳鲹肠道组织和肝脏组织中的表达差异(图8),结果显示,在肠道组织中ToKiss1基因mRNA在喂食颗粒饲料的组中表达量最高,其次为喂食杂鱼组,在喂食鱿鱼组中表达量最低(P<0.05);在肝脏组织中,3组之间均无显著差异(P>0.05)。
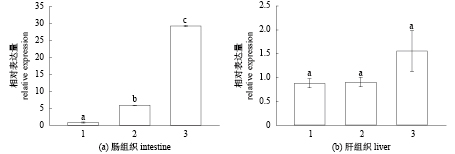
|
图 8 不同实验组卵形鲳鲹ToKiss1 mRNA的表达 1. 冰鱿鱼组,2. 冰杂鱼组,3. 颗粒饲料组 Fig. 8 The expression level of ToKiss1 in different test groups 1. iced squid group, 2. iced fish group, 3. pelleted feed group |
Kisspeptin系统对硬骨鱼的生长和能量消耗具有重要的调控作用。根据保守核心序列区氨基酸的数目不同将脊椎动物kisspeptin成熟肽划分为6种类型,即kisspeptin-54、−15、−14、−13、−12、−10,在鱼中仅发现kisspeptin-10类型[29]。本研究首次成功克隆了卵形鲳鲹ToKiss1基因,ToKiss1基因具有核心序列“YNLNSFGLRY”,与其他硬骨鱼如青鳉[9]、斑马鱼[10]、日本鲭[15]和花鲈[5]的kisspeptin-10核心序列相同,所以ToKiss1氨基酸属于kisspeptin-10类型。哺乳动物kisspeptin-10的核心序列为“YNWNSFGLRY”,仅有第3位氨基酸与硬骨鱼不同,由此可见Kiss1基因的核心功能中心在进化中高度保守。
kisspeptin作为神经多肽在脑中广泛表达,硬骨鱼类Kiss1基因主要在松果体(habenula)和视前叶—下丘脑(preoptic-hypothalamic)中表达[30]。博纳里牙汉鱼(Odontesthes bonariensis)成鱼Kiss1基因mRNA表达于大脑的各个部分[31]。抚仙金线鲃[32]的Kiss1基因mRNA在脑中广泛表达(下丘脑、垂体、脑),在垂体和下丘脑中的表达量较高。斑马鱼的Kiss1 mRNA在间脑和中脑高表达,在后脑中等表达,在端脑和垂体低表达,在胰和肠前部也有表达[33]。ToKiss1基因mRNA在正常卵形鲳鲹的脑组织中具有较高的表达量,与其他硬骨鱼类相似。Kiss1基因mRNA在与繁殖无关的组织和器官中也具有广泛的分布。金鱼Kiss1在视顶盖—丘脑、肠道、肾脏和精巢等中高度表达[14],Yang等[34]研究发现其在脂肪、鳃和肝脏中也有表达,且在肠道中表达量最高。斑马鱼的Kiss1基因mRNA在胰腺和肠道前部也有表达[33],抚仙金线鲃Kiss1基因mRNA在肠道的表达量最高。该研究中卵形鲳鲹ToKiss1基因具有相似的表达组织,在肠道、胃、脾脏、肌肉、心脏等组织中均有表达。目前关于kisspeptin的功能研究主要集中在生殖内分泌调控方面,但Kiss1基因mRNA在肠道、胃等组织中的分布显示kisspeptin还可能与消化系统功能相关。ToKiss1在肌肉和脾脏中的分布,显示其可能参与鱼体的生理代谢,和免疫等功能相关。鱼类Kiss1系统的研究主要局限在参与鱼体的性腺轴调控等方面。但在哺乳类中已有发现,Kiss1系统参与调控能量平衡[24]。小鼠下丘脑Kiss1基因的表达均受到维持机体葡萄糖稳态的胰岛素和机体代谢相关激素LP、NPY的影响[20,22]。小鼠脂肪组织中Kiss1基因在短期禁食后表达量上升,以促进脂肪组织中能量形式的转变[21]。在解剖学上kisspeptin神经元与厌食症的POMC神经元相连,并能直接激活或间接抑制促进食欲的NPY神经元。由此可见,kisspeptin可能在调节能量平衡中发挥直接作用[35]。
肠道和胰腺内的分泌细胞可分泌大量对肠道运动、肠道分泌和食欲都有影响的多肽,如胃泌酸调节素(oxyntomodulin)被证明可以减少体质量和食物摄入量;主食激素促分泌素受体的内源性配体(ghrelin)被证明可以增加食欲和体质量;肥胖抑制素(obestatin)被证实是一种新的肠肽;YY肽(3-36)被证明具有厌食活性[36]。该研究中ToKiss1基因在卵形鲳鲹肠道组织中的表达量仅次于脑组织,饵料投喂的卵形鲳鲹ToKiss1 基因mRNA在肠道组织中的表达分析结果表明,饵料类型显著影响ToKiss1基因在肠道组织中的表达。Sun等[28]发现,颗粒饲料组的LC-PUFA含量最高且生长最快,其次是冰杂鱼组,冰鱿鱼组最低。由此可见,饵料类型可能影响ToKiss1基因在肠道组织中的表达来促进摄食和食物消化吸收,进而影响卵形鲳鲹的个体生长。高脂饮食通常导致养殖鱼类肝脏中脂肪堆积过多,同时伴随着生长缓慢[37-38]。肝脏是脂肪代谢的重要场所,调控鱼类摄食、葡萄糖和脂肪代谢的重要激素LP被证实主要合成部位在鱼类肝脏[39];最能有效促进食欲的NPY在肝脏中也有分布[40];Bergan等[41]报道在虹鳟(Oncorhynchus mykiss)离体培养的肝细胞中,GH可以通过激活激素敏感脂肪酶(hormone-sensitive lipase, HSL)来促进脂肪分解。该研究中ToKiss1基因在卵形鲳鲹肝脏组织中虽有分布,但3种不同饵料对卵形鲳鲹ToKiss1基因mRNA在肝组织中的表达无显著影响。由此推测ToKiss1可能在肝脏中不直接参与调节脂代谢。
| [1] |
Lee J H, Miele M E, Hicks D J, et al. Kiss-1, a novel human malignant melanoma metastasis-suppressor gene[J].
Journal of the National Cancer Institute, 1996, 88(23): 1731-1737 DOI: 10.1093/jnci/88.23.1731 ( 0) 0)
|
| [2] |
Kanda S, Karigo T, Oka Y. Steroid sensitive kiss2 neurones in the goldfish: evolutionary insights into the duplicate kisspeptin gene-expressing neurones
[J].
Journal of Neuroendocrinology, 2012, 24(6): 897-906 DOI: 10.1111/j.1365-2826.2012.02296.x ( 0) 0)
|
| [3] |
Shahjahan M, Motohashi E, Doi H, et al. Elevation of Kiss2 and its receptor gene expression in the brain and pituitary of grass puffer during the spawning season[J].
General and Comparative Endocrinology, 2010, 169(1): 48-57 DOI: 10.1016/j.ygcen.2010.07.008 ( 0) 0)
|
| [4] |
Mechaly A S, Viñas J, Piferrer F. Gene structure analysis of kisspeptin-2 (Kiss2) in the Senegalese sole (Solea senegalensis): characterization of two splice variants of Kiss2, and novel evidence for metabolic regulation of kisspeptin signaling in non-mammalian species
[J].
Molecular and Cellular Endocrinology, 2011, 339(1-2): 14-24 DOI: 10.1016/j.mce.2011.03.004 ( 0) 0)
|
| [5] |
Felip A, Zanuy S, Pineda R, et al. Evidence for two distinct KiSS genes in non-placental vertebrates that encode kisspeptins with different gonadotropin-releasing activities in fish and mammals
[J].
Molecular and Cellular Endocrinology, 2009, 312(1-2): 61-71 DOI: 10.1016/j.mce.2008.11.017 ( 0) 0)
|
| [6] |
Force A, Lynch M, Pickett F B, et al. Preservation of duplicate genes by complementary, degenerative mutations[J].
Genetics, 1999, 151(4): 1531-1545 ( 0) 0)
|
| [7] |
Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization[J].
Genetics, 2000, 154(1): 459-473 ( 0) 0)
|
| [8] |
Nakatani Y, Takeda H, Kohara Y, et al. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates[J].
Genome Research, 2007, 17(9): 1254-1265 DOI: 10.1101/gr.6316407 ( 0) 0)
|
| [9] |
Kanda S, Akazome Y, Matsunaga T, et al. Identification of Kiss-1 product kisspeptin and steroid-sensitive sexually dimorphic kisspeptin neurons in medaka (Oryzias latipes)
[J].
Endocrinology, 2008, 149(5): 2467-2476 DOI: 10.1210/en.2007-1503 ( 0) 0)
|
| [10] |
Kitahashi T, Ogawa S, Parhar I S, et al. Cloning and Expression of kiss2 in the zebrafish and medaka
[J].
Endocrinology, 2009, 150(2): 821-831 DOI: 10.1210/en.2008-0940 ( 0) 0)
|
| [11] |
van Aerle R, Kille P, Lange A, et al. Evidence for the existence of a functional Kiss1/Kiss1 receptor pathway in fish[J].
Peptides, 2008, 29(1): 57-64 DOI: 10.1016/j.peptides.2007.10.018 ( 0) 0)
|
| [12] |
Migaud H, Ismail R, Cowan M, et al. Kisspeptin and seasonal control of reproduction in male European sea bass (Dicentrarchus labrax)
[J].
General and Comparative Endocrinology, 2012, 179(3): 384-399 DOI: 10.1016/j.ygcen.2012.07.033 ( 0) 0)
|
| [13] |
Servili A, Le Page Y, Leprince J, et al. Organization of two independent kisspeptin systems derived from evolutionary-ancient Kiss genes in the brain of zebrafish
[J].
Endocrinology, 2011, 152(4): 1527-1540 DOI: 10.1210/en.2010-0948 ( 0) 0)
|
| [14] |
Li S S, Zhang Y, Liu Y, et al. Structural and functional multiplicity of the kisspeptin/GPR54 system in goldfish (Carassius auratus)
[J].
Journal of Endocrinology, 2009, 201(3): 407-418 DOI: 10.1677/JOE-09-0016 ( 0) 0)
|
| [15] |
Selvaraj S, Kitano H, Fujinaga Y, et al. Molecular characterization, tissue distribution, and mRNA expression profiles of two Kiss genes in the adult male and female chub mackerel (Scomber japonicus) during different gonadal stages
[J].
General and Comparative Endocrinology, 2010, 169(1): 28-38 DOI: 10.1016/j.ygcen.2010.07.011 ( 0) 0)
|
| [16] |
Zmora N, Stubblefield J, Zulperi Z, et al. Differential and gonad stage-dependent roles of Kisspeptin1 and Kisspeptin2 in reproduction in the modern Teleosts, Morone species
[J].
Biology of Reproduction, 2012, 86(6): 177 DOI: 10.1095/biolreprod.111.097667 ( 0) 0)
|
| [17] |
Shi Y, Zhang Y, Li S S, et al. Molecular identification of the Kiss2/Kiss1ra system and its potential function during 17Alpha-Methyltestosterone-Induced sex reversal in the orange-spotted grouper, Epinephelus coioides
[J].
Biology of Reproduction, 2010, 83(1): 63-74 DOI: 10.1095/biolreprod.109.080044 ( 0) 0)
|
| [18] |
Mechaly A S, Viñas J, Piferrer F. Sex-specific changes in the expression of kisspeptin, kisspeptin receptor, gonadotropins and gonadotropin receptors in the Senegalese sole (Solea senegalensis) during a full reproductive cycle
[J].
Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 2012, 162(4): 364-371 DOI: 10.1016/j.cbpa.2012.04.003 ( 0) 0)
|
| [19] |
Alvarado M V, Carrillo M, Felip A. Expression of kisspeptins and their Receptors, gnrh-1/gnrhr-II-1a and gonadotropin genes in the brain of adult male and female European sea bass during different gonadal stages[J].
General and Comparative Endocrinology, 2013, 187: 104-116 DOI: 10.1016/j.ygcen.2013.03.030 ( 0) 0)
|
| [20] |
Qiu X L, Dowling A R, Marino J S, et al. Delayed puberty but normal fertility in mice with selective deletion of insulin receptors from Kiss1 cells
[J].
Endocrinology, 2013, 154(3): 1337-1348 DOI: 10.1210/en.2012-2056 ( 0) 0)
|
| [21] |
Brown R E, Imran S A, Ur E, et al. Kiss-1 mRNA in adipose tissue is regulated by sex hormones and food intake
[J].
Molecular and Cellular Endocrinology, 2008, 281(1-2): 64-72 DOI: 10.1016/j.mce.2007.10.011 ( 0) 0)
|
| [22] |
Luque R M, Kineman R D, Tena-Sempere M. Regulation of hypothalamic expression of Kiss-1 and GPR54 genes by metabolic factors: analyses using mouse models and a cell line[J].
Endocrinology, 2007, 148(10): 4601-4611 DOI: 10.1210/en.2007-0500 ( 0) 0)
|
| [23] |
Jiang Q, He M L, Ko W K W, et al. Kisspeptin induction of somatolactin-α release in goldfish pituitary cells: functional role of cAMP/PKA-, PLC/PKC-, and Ca2+/calmodulin-dependent cascades
[J].
American Journal of Physiology-Endocrinology and Metabolism, 2014, 307(10): E872-E884 DOI: 10.1152/ajpendo.00321.2014 ( 0) 0)
|
| [24] |
Tolson K P, Garcia C J, Smith J T, et al. Kisspeptin signaling is a novel player in obesity, metabolism, and glucose homeostasis in female mice[C]//Endocrine Society’s 96th Annual Meeting and Expo. Chicago: Endocrine Society, 2014.
( 0) 0)
|
| [25] |
Cheng Q T, Zheng B S. Systematic Synopsis of Chinese Fishes[M]. Beijing: Science Press, 1987: 341-342(in Chinese).
( 0) 0)
|
| [26] |
Xie Z Z, Xiao L, Wang D D, et al. Transcriptome analysis of the Trachinotus ovatus: identification of reproduction, growth and immune-related genes and microsatellite markers
[J].
PLoS One, 2014, 9(10): e109419 DOI: 10.1371/journal.pone.0109419 ( 0) 0)
|
| [27] |
Tutman P, Glavić N, Kožul V, et al. Preliminary information on feeding and growth of pompano, Trachinotus ovatus (Linnaeus, 1758) (pisces; carangidae) in captivity
[J].
Aquaculture International, 2004, 12(4-5): 387-393 DOI: 10.1023/B:AQUI.0000042135.88381.f4 ( 0) 0)
|
| [28] |
Sun L Y, Zhang D C, Guo H Y, et al. Complete mitochondrial genome sequence of golden pompano Trachinotus ovatus
[J].
Mitochondrial DNA Part A, 2016, 27(2): 871-872 DOI: 10.3109/19401736.2014.919482 ( 0) 0)
|
| [29] |
Akazome Y, Kanda S, Okubo K, et al. Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain[J].
Journal of Fish Biology, 2010, 76(1): 161-182 DOI: 10.1111/j.1095-8649.2009.02496.x ( 0) 0)
|
| [30] |
Ogawa S, Parhar I S. Anatomy of the kisspeptin systems in teleosts[J].
General and Comparative Endocrinology, 2013, 181: 169-174 DOI: 10.1016/j.ygcen.2012.08.023 ( 0) 0)
|
| [31] |
Bohórquez M O T, Mechaly A S, Hughes L C, et al. Kisspeptin system in pejerrey fish (Odontesthes bonariensis): characterization and gene expression pattern during early developmental stages
[J].
Comparative Biochemistry and Physiology-Part A: Molecular & Integrative Physiology, 2017, 204: 146-156 DOI: 10.1016/j.cbpa.2016.11.014 ( 0) 0)
|
| [32] |
廖宗甄, 孙彩云, 王晓爱, 等. 抚仙金线鲃kisspeptin基因的克隆及组织表达特性[J].
四川动物, 2016, 35(4): 496-502 Liao Z Z, Sun C Y, Wang X A, et al. Molecular cloning and tissue distribution of kisspeptin from Sinocyclocheilus tingi [J]. Sichuan Journal of Zoology, 2016, 35(4): 496-502 (  0) 0)
|
| [33] |
Biran J, Ben-Dor S, Levavi-Sivan B. Molecular identification and functional characterization of the kisspeptin/kisspeptin receptor system in lower vertebrates[J].
Biology of Reproduction, 2008, 79(4): 776-786 DOI: 10.1095/biolreprod.107.066266 ( 0) 0)
|
| [34] |
Yang B, Jiang Q, Chan T, et al. Goldfish kisspeptin: molecular cloning, tissue distribution of transcript expression, and stimulatory effects on prolactin, growth hormone and luteinizing hormone secretion and gene expression via direct actions at the pituitary level[J].
General and Comparative Endocrinology, 2010, 165(1): 60-71 DOI: 10.1016/j.ygcen.2009.06.001 ( 0) 0)
|
| [35] |
de Bond J A, Smith J T. Kisspeptin and energy balance in reproduction[J].
Reproduction, 2014, 147(3): R53-R63 DOI: 10.1530/REP-13-0509 ( 0) 0)
|
| [36] |
Baynes K C, Dhillo W S, Bloom S R. Regulation of food intake by gastrointestinal hormones[J].
Current Opinion in Gastroenterology, 2006, 22(6): 626-631 DOI: 10.1097/01.mog.0000245537.43142.63 ( 0) 0)
|
| [37] |
Regost C, Arzel J, Cardinal M, et al. Dietary lipid level, hepatic lipogenesis and flesh quality in turbot (Psetta maxima)
[J].
Aquaculture, 2001, 193(3-4): 291-309 DOI: 10.1016/S0044-8486(00)00493-2 ( 0) 0)
|
| [38] |
Wang Z L, Chua H K, Gusti A A R A, et al. RING-H2 protein WSSV249 from white spot syndrome virus sequesters a shrimp ubiquitin-conjugating enzyme, PvUbc, for viral pathogenesis[J].
Journal of Virology, 2005, 79(14): 8764-8772 DOI: 10.1128/JVI.79.14.8764-8772.2005 ( 0) 0)
|
| [39] |
卢荣华, 孙君君, 梁旭方, 等. 鱼类leptin的生物学特性及功能[J].
水生生物学报, 2015, 39(3): 583-589 Lu R H, Sun J J, Liang X F, et al. Biological characteristics and functions of leptin in fish[J]. Acta Hydrobiologica Sinica, 2015, 39(3): 583-589 (  0) 0)
|
| [40] |
Yang Z Y. Effect of fasting-refeeding on neuropeptide Y and leptins in silver carp (Hypophthalmichthys molitrix)[D]. Wuhan: Huazhong Agricultural University, 2015(in Chinese).
( 0) 0)
|
| [41] |
Bergan H E, Kittilson J D, Sheridan M A. PKC and ERK mediate GH-stimulated lipolysis[J].
Journal of Molecular Endocrinology, 2013, 51(2): 213-224 DOI: 10.1530/JME-13-0039 ( 0) 0)
|
2. College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306, China;
3. Guangdong Provincial Engineer Technology Research Center of Marine Biological Seed Industry, Guangzhou 510300, China;
4. Guangdong Haid Group Co., Ltd., Guangzhou 511400, China
 2019, Vol. 43
2019, Vol. 43


